Introduction:
More than half of human genes undergo alternative polyadenylation (APA) and generate mRNA
transcripts with varying lengths. Increasing awareness of APA’s role in human health and disease
has propelled the development of several 3’ sequencing (3’Seq) techniques. Despite the recent
data explosion, computational tools that are precisely designed for 3’Seq data are not well
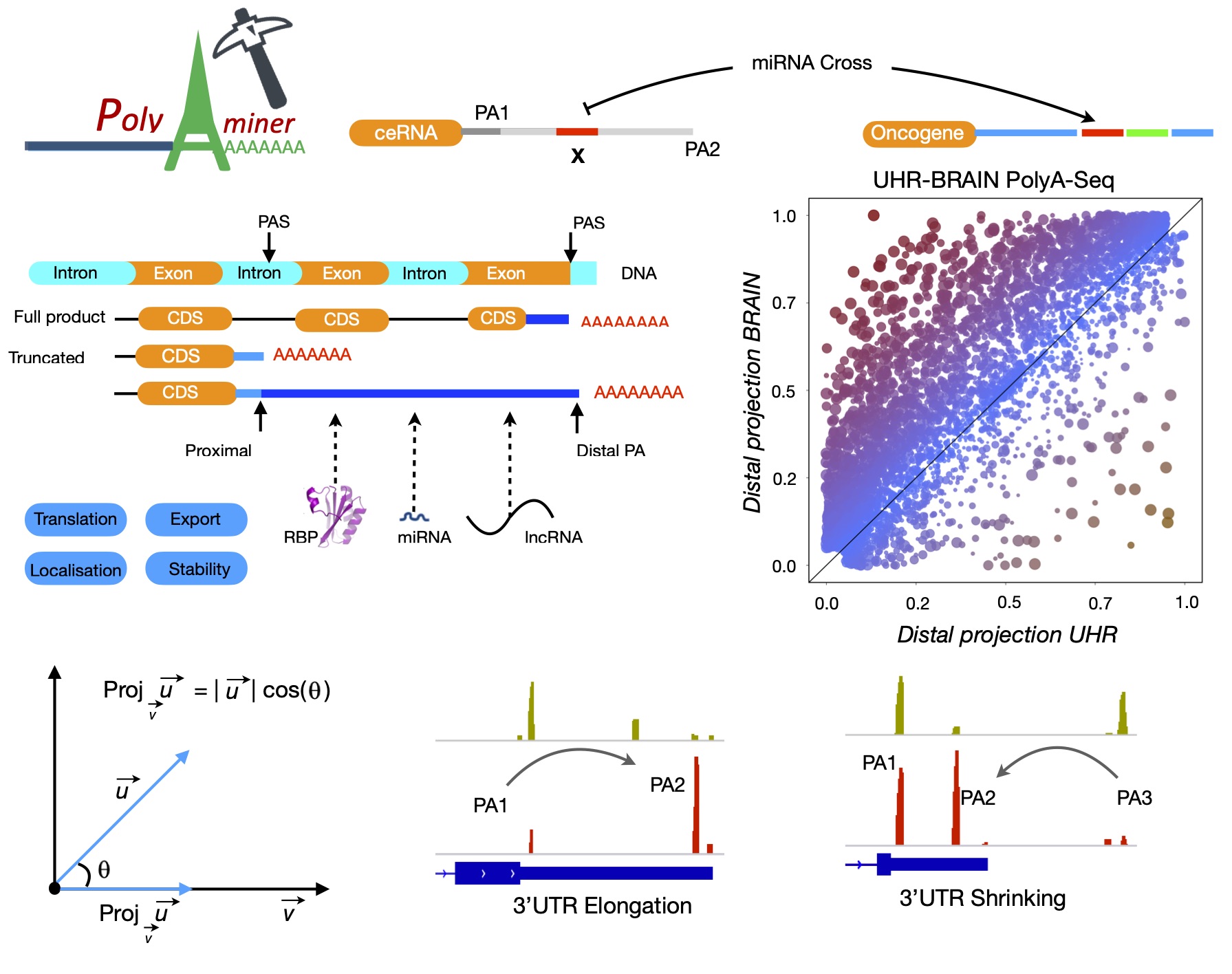
established. PolyA-miner is developed specifically for 3’Seq data, it accounts for all
non‐proximal to non‐distal APA switches using vector projections and reflect precise gene level
3’UTR changes. PolyA-miner is less susceptible to inherent data variations can also to
effectively identify novel APA sites that are otherwise undetected using reference-based
approaches. With the emerging importance of alternative polyadenylation in studying human
diseases, PolyA-miner can significantly accelerate data analysis and help decoding the missing
pieces of underlying alternative polyadenylation dynamics.